application note sur l'amplicon deep sequencing avec illumina miseq/nextseq
guide technique sur l'amplicon deep sequencing utilisant illumina
Contenu du document
the swiss dna company
application note · next generation sequencing
amplicon deep sequencing on the illumina miseq/nextseq platform
- detect introduced or expected mutations
- discover rare genomic variations with high confidence
introduction
amplicon deep sequencing using next generation sequencing (NGS) technologies has become a powerful tool to study a wide variety of research questions. typical applications include:
- CRISPR genome editing protocols of eukaryotes
- genome-wide transposon insertion analysis in microorganisms
- human leukocyte antigen (HLA) typing
- screening of specific somatic mutations in tumor tissues
a commonality among these approaches is that a PCR is used to amplify fragments, which are then sequenced on a single-molecule level.
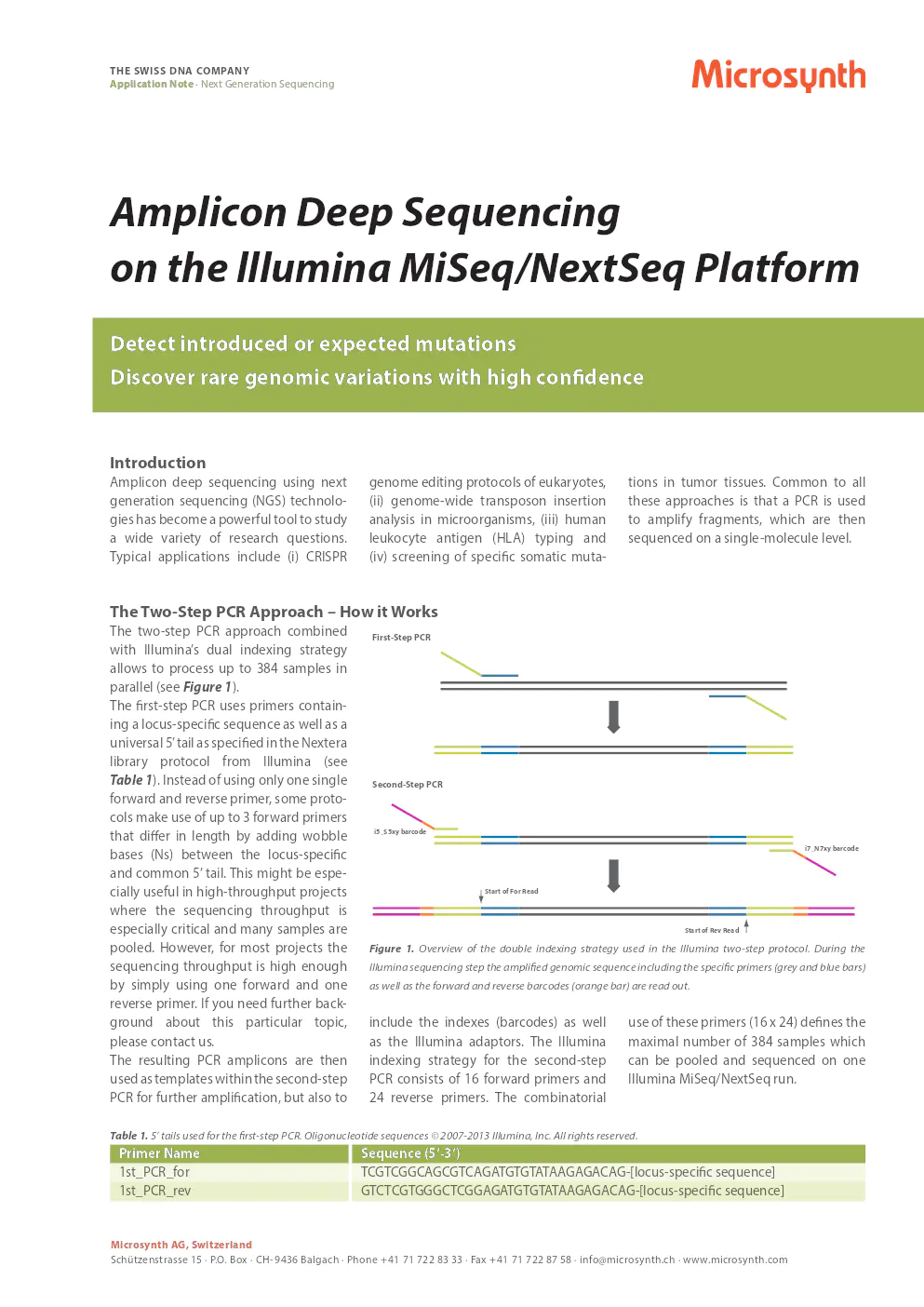
the two-step PCR approach – how it works
the two-step PCR approach, combined with illumina’s dual indexing strategy, allows processing of up to 384 samples in parallel. the first-step PCR uses primers containing a locus-specific sequence as well as a universal 5’ tail as specified in the nextera library protocol from illumina.
first-step PCR
instead of using only one single forward and reverse primer, some protocols use up to 3 forward primers that differ in length by adding wobble bases (Ns) between the locus-specific and common 5’ tail. this is especially useful in high-throughput projects where sequencing throughput is critical and many samples are pooled. for most projects, high enough sequencing throughput can be achieved with a single forward and reverse primer. for further background, contact us.
second-step PCR
the resulting PCR amplicons serve as templates within the second-step PCR for further amplification, and to include indexes (barcodes) and illumina adaptors. illumina indexing for the second-step PCR consists of 16 forward primers and 24 reverse primers. this combinatorial use allows pooling and sequencing up to 384 samples in one illumina miseq/nextseq run.
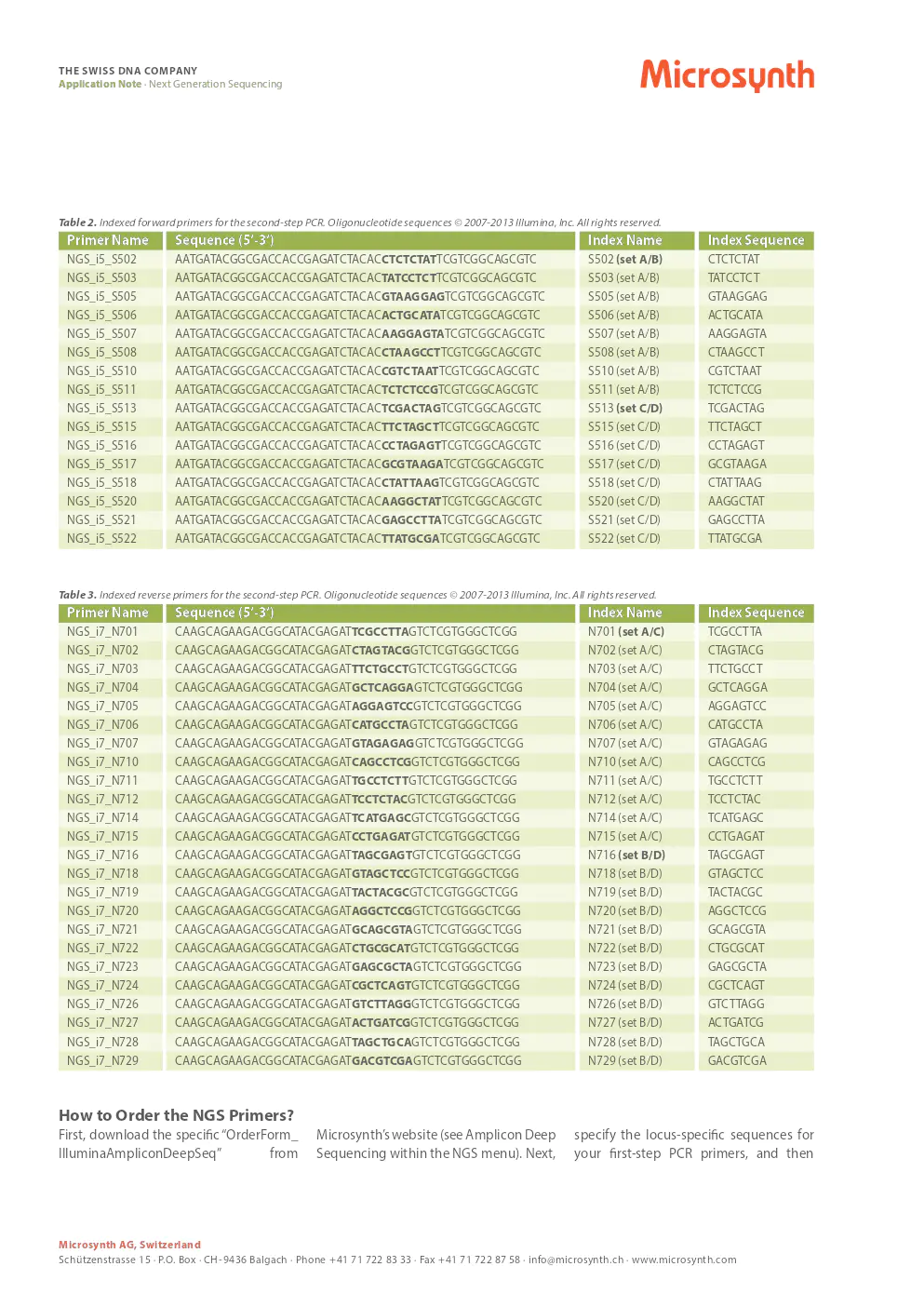
table 1. 5’ tails used for the first-step PCR.
primer name sequence (5‘-3‘)
- 1st_PCR_for: tcgtcggcagcgtcagatgtgtataagagacag-[locus-specific sequence]
- 1st_PCR_rev: gtctcgtgggctcggagatgtgtataagagacag-[locus-specific sequence]
microsynth ag, switzerland
schützenstrasse 15 · P.O. box · CH?-?9436 balgach · phone +?41 71 722 83?33 · fax +?41 71 722 87?58 · info@microsynth.ch · www.microsynth.com
how to order the NGS primers?
first, download the specific “orderform_illuminaamplicondeepseq” from microsynth’s website from the amplicon deep sequencing section within the NGS menu. specify the locus-specific sequences for your first-step PCR primers, and then select your desired indexed forward and reverse primers. send the upload sheet to info@microsynth.ch and request your specific offer. alternatively, directly order the oligos in our webshop using the prefix “NGS_” in the 0.2 ?mol scale, HPLC purified.
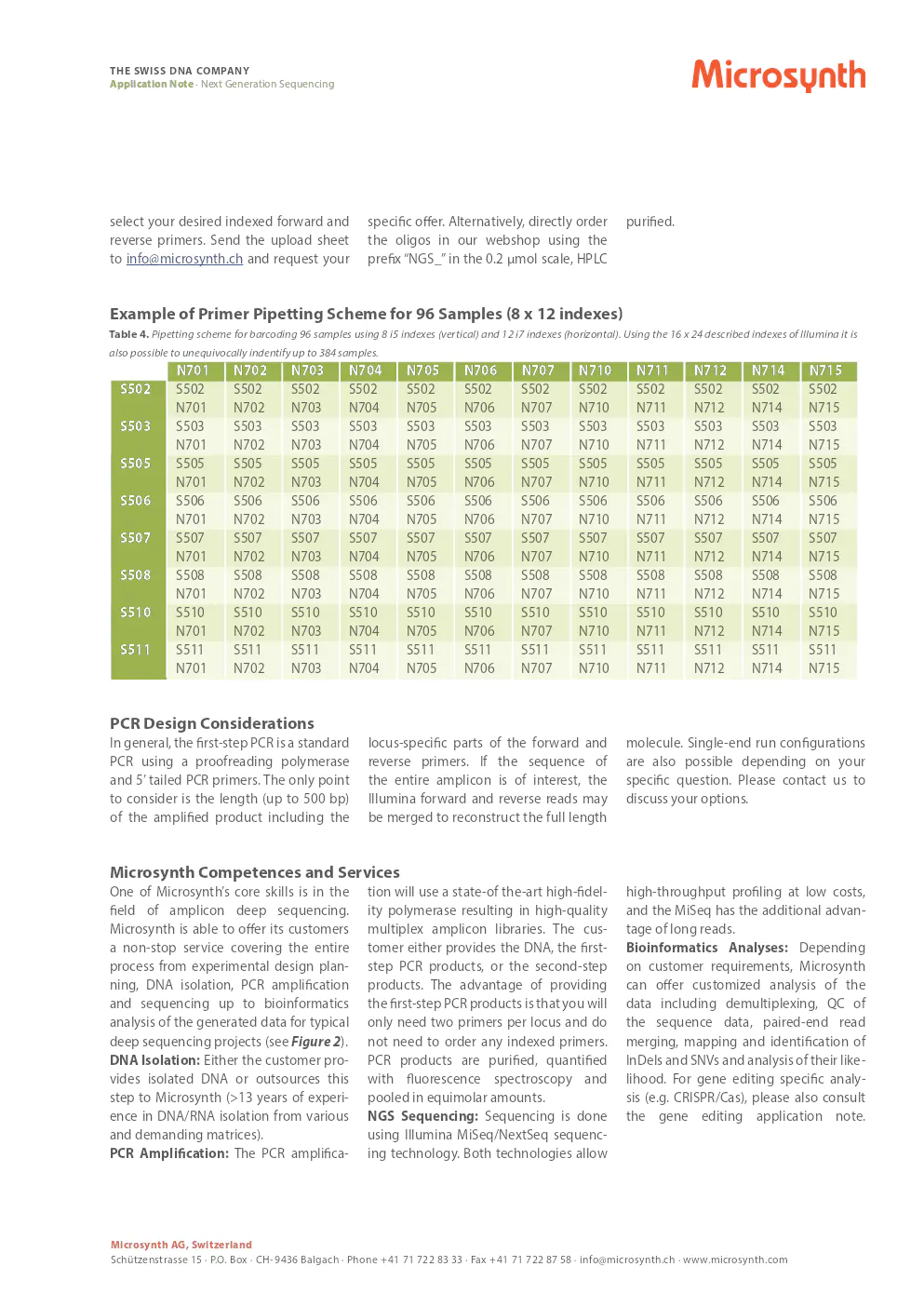
example of primer pipetting scheme for 96 samples
table 4. pipetting scheme for barcoding 96 samples using 8 i5 indexes (vertical) and 12 i7 indexes (horizontal). using the 16 x 24 described indexes of illumina, it is also possible to unequivocally identify up to 384 samples.
PCR design considerations
in general, the first-step PCR is a standard PCR using a proofreading polymerase and 5’ tailed PCR primers. consider the length (up to 500 bp) of the amplified product including locus-specific parts of the forward and reverse primers. if the sequence of the entire amplicon is of interest, illumina forward and reverse reads may be merged to reconstruct the full-length molecule. single-end run configurations are also possible depending on your specific question. please contact us to discuss options.
microsynth competences and services
microsynth specializes in amplicon deep sequencing, offering a comprehensive service from experimental design, DNA isolation, PCR amplification, sequencing, to bioinformatics analysis of the generated data.
DNA isolation: microsynth provides a DNA isolation service, utilizing over 13 years of experience in DNA/RNA isolation from diverse matrices.
PCR amplification: using a high-fidelity polymerase, microsynth can generate high-quality multiplex amplicon libraries.
NGS sequencing: sequencing is done using illumina miseq/nextseq technology, supporting high-throughput profiling at low costs. miseq offers the advantage of long reads.
bioinformatics analyses: customized analysis options include demultiplexing, QC, paired-end read merging, mapping, and identification of indels and SNVs. consult the gene editing application note for CRISPR/Cas specific analyses.
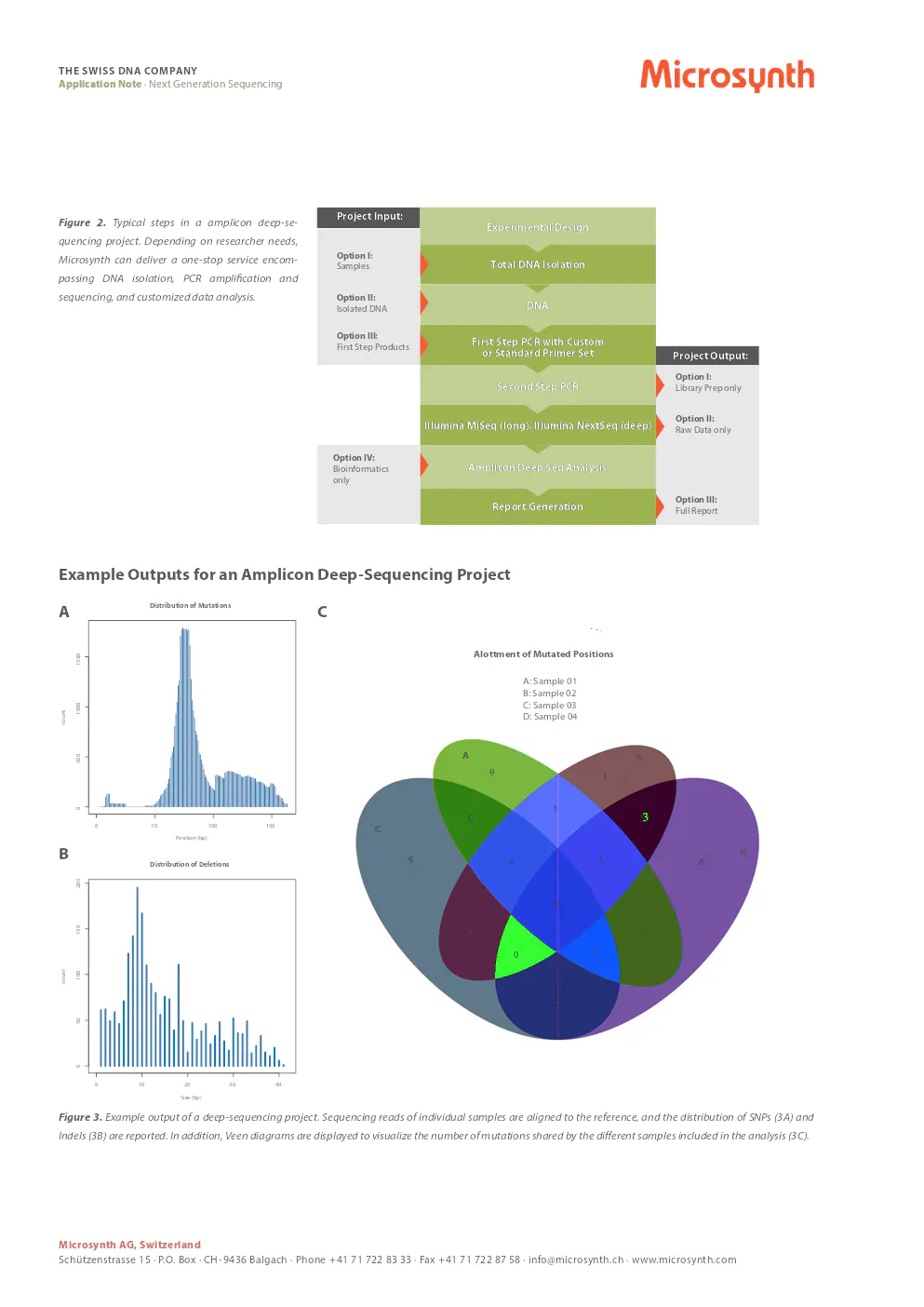
example outputs for an amplicon deep-sequencing project
figure 3. example output of a deep-sequencing project. sequencing reads of samples are aligned to a reference, and distributions of SNPs and indels are reported. veen diagrams visualize mutations shared by included samples.
microsynth ag, switzerland
schützenstrasse 15 · P.O. box · CH?-?9436 balgach · phone +?41 71 722 83?33 · fax +?41 71 722 87?58 · info@microsynth.ch · www.microsynth.com